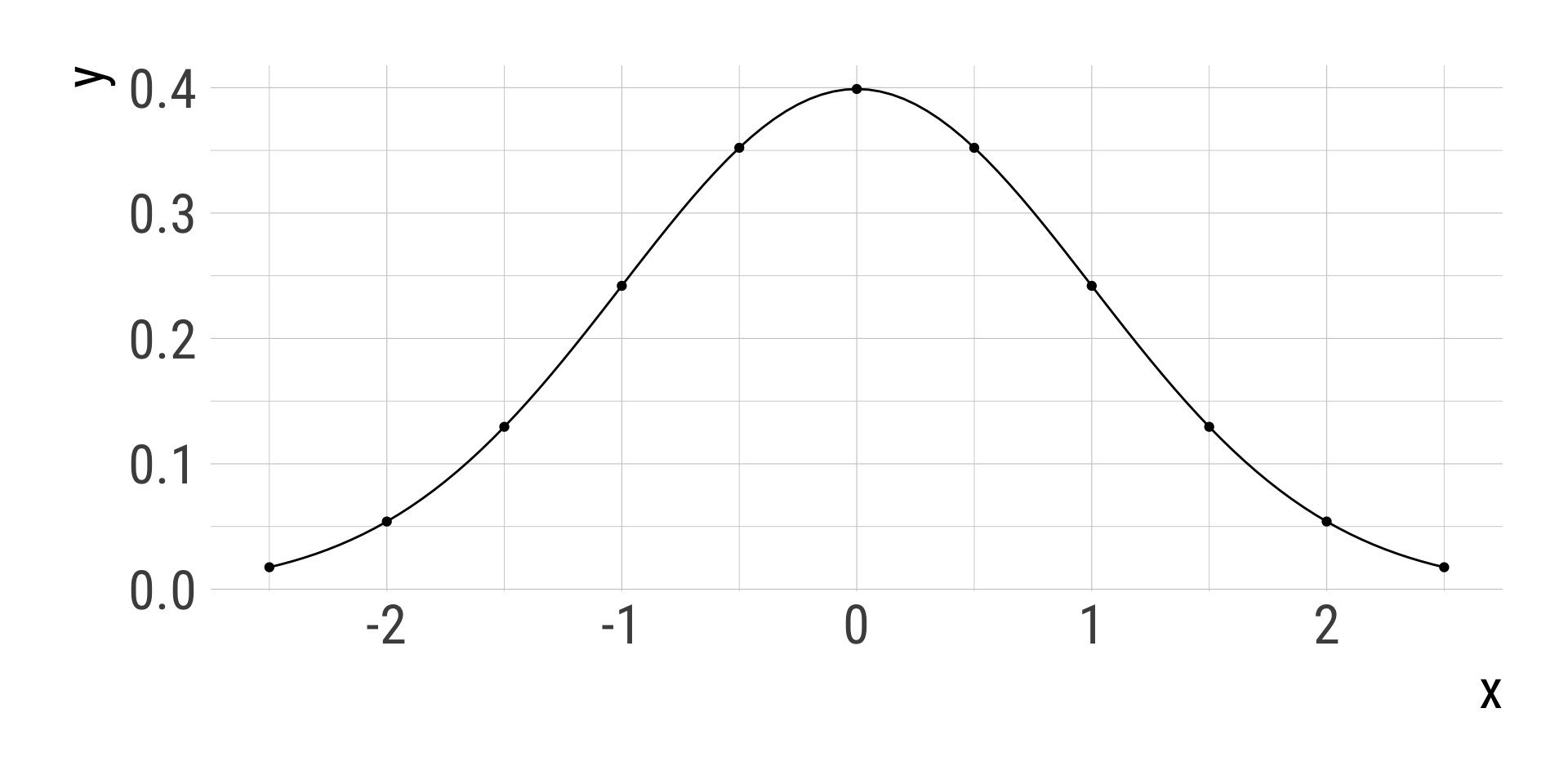
dnorm(x = 0, mean = 0, sd = 1, log = FALSE)[1] 0.3989423dnorm(x = seq(-2.5, 2.5, by = 0.5), mean = 0, sd = 1, log = FALSE) [1] 0.01752830 0.05399097 0.12951760 0.24197072 0.35206533 0.39894228
[7] 0.35206533 0.24197072 0.12951760 0.05399097 0.01752830